Deployment Options¶
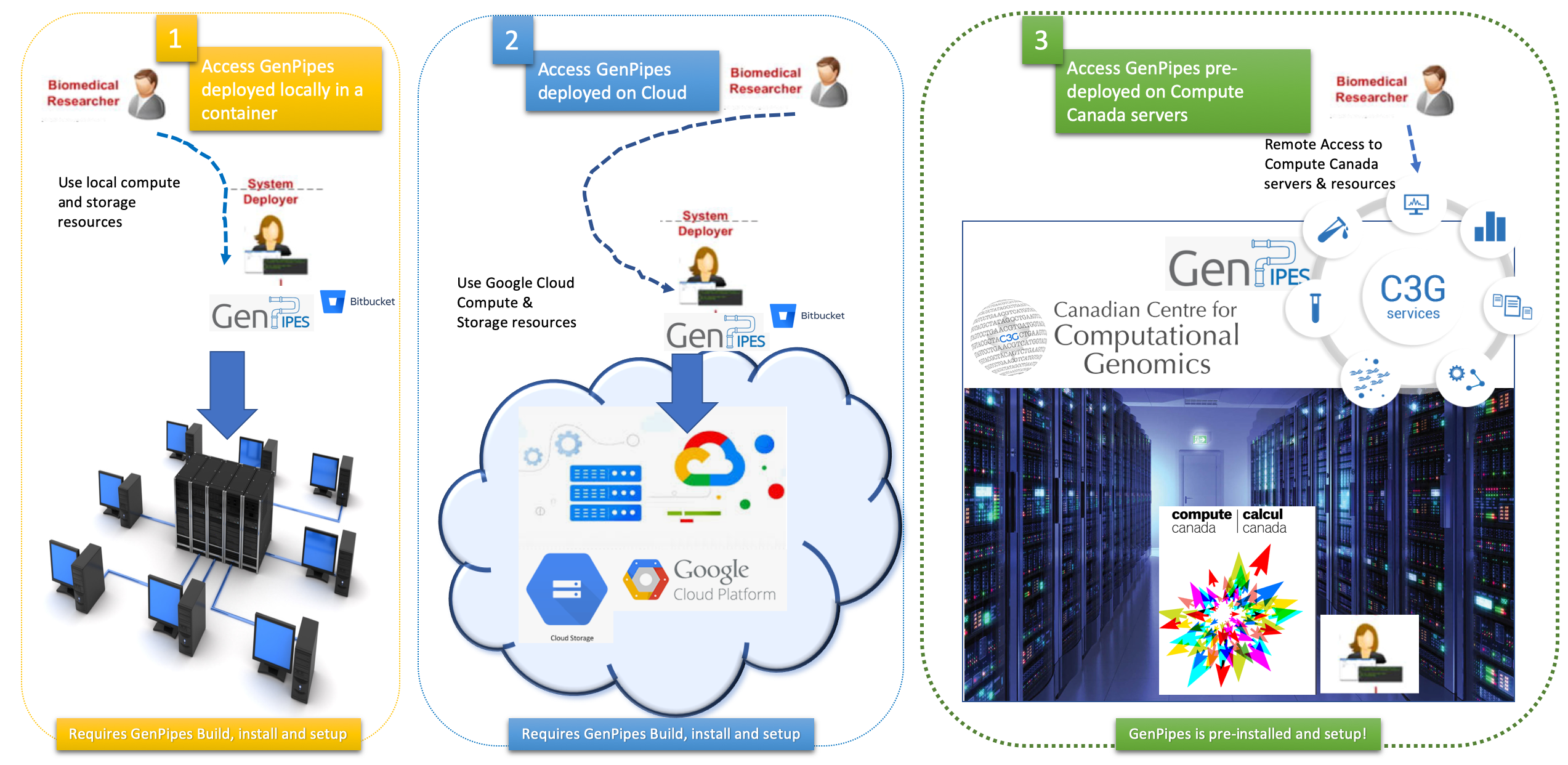
There are multiple ways to access GenPipes and get started with genomic analysis using the pipelines therein. Figure below represents the three options available to bioinformatics researchers to access GenPipes.
Remotely access GenPipes deployed at Compute Canada infrastructure
GenPipes deployment in the cloud - Google Cloud Platform (GCP)
Local deployment
Bare metal or virtual server
GenPipes in a container
Accessing GenPipes¶
The infographic below represents various mechanisms that can be used to access GenPipes today.
Obtaining GenPipes sources¶
Refer to the latest GenPipes sources for instructions on downloading and setting up GenPipes.
GenPipes on Compute Canada infrastructure¶
Researchers who have access to Compute Canada resources need not deploy GenPipes for genomic analysis. They can simply login and access Compute Canada servers that have pre-installed stable release of GenPipes. For details refer to Accessing GenPipes deployment on Compute Canada infrastructure. External users who do not have access to Compute Canada data centre resources can apply for the same.
Through a partnership with the Compute Canada consortium, the pipelines and third-party tools have also been configured on 6 different Compute Canada HPC centers. This allows any Canadian researcher to use GenPipes along with the needed computing resources by simply applying to the consortium. To ensure consistency of pipeline versions and used dependencies (such as genome references and annotation files) and to avoid discrepancy between compute sites, pipeline set-up has been centralized to 1 location, which is then distributed on a real-time shared file system: the CERN (European Organization for Nuclear Research) Virtual Machine File System CVM FS.
GenPipes deployment on GCP¶
If you need to run large scale genomic analysis that requires resource scaling, GenPipes can be deployed and accessed from cloud. At present, Google Compute Platform (GCP) is supported. You may require assistance from System Administrator or your local cloud expert to install and deploy GenPipes in the cloud before you can access and run genomic analysis pipelines provided by GenPipes. For details refer to GenPipes installation guide section titled “How to deploy GenPipes in the cloud?”.
Local deployment¶
If you wish to deploy GenPipes locally using your own compute and storage infrastructure, you can refer to the BitBucket repository listed earlier. You could either deploy it on your local server / workstation or try GenPipes in a container option.
GenPipes can be installed from scratch on any Linux cluster supporting Python 2.7 by following the instructions in the README.md file. GenPipes can also be deployed via containers approach. A Docker image of GenPipes is available which simplifies the set-up process and can be used on a range of platforms, including cloud platforms. This allows system-wide installations, as well as local user installations via the Docker image without needing special permissions.
Local deployment option can be used for small scale genomic analyses using genome datasets available locally. The GenPipes in a container option is a self-contained image that offers GenPipes software, common reference genomes and all that is needed to run the pre-built analysis pipelines. Bioinformatics researchers who are not familiar with container technology may require assistance from System Administrators in deploying a local copy of GenPipes software. For details refer to GenPipes installation guide section for container deployment GenPipes in a container.